Antimicrobial resistance (AMR)
Antimicrobial resistance (AMR)
“The future of humanity and microbes will likely evolve as episodes of our wits versus their genes. Their genes allow them to adapt to anything we through at them. It is up to us to use our wits to keep up with their genes” – Nobel Laureate, Dr Joshua Lederberg.
Antimicrobial resistance (AMR) is a phenomenon where the previously wonder drugs called antibiotics becomes ineffective to treat formerly simpler infection during post antibiotic discovery era. As Dr. Lederberg rightly said “It is upto us to use our wits to keep up with their genes”. In fact the first instance of AMR was observed at the similar time when the first antibiotic was deemed to be used for human welfare. AMR is rightly threatening the effective use of antimicrobials against various infection caused by pathogenic microbes. WHO recognized it as a global problem way back in 2001. Pathogenic microbes constitutively including bacteria, virus, fungi, parasites etc. where antimicrobials such antiviral, antibiotics, antifungals were largely effective to eradicate their infections are becoming resistant. Microbes who exhibit AMR are sometimes referred as “Superbugs”. Prediction regarding AMR by leading ex-Goldman Sachs economist Lord Jim O’Neill – writes that there has been encouraging progress in some areas such as a raised profile for AMR, funding for early-stage antibiotics research and the number of scientists working in this area. If neglected the world may see 10 million deaths due to AMR per year by 2050 with a cumulative loss of $100 trillion to the global economy.
Evolution of AMR can occur naturally or through environmental pressure in an antibiotic laden environment where a few survive and reproduce to form a fully AMR colony. The common mechanism by which a microbe exhibit AMR are: drug inactivation or modification, alteration of target site, alteration of metabolic pathway, reduced drug accumulation and increased active efflux of the drugs across the cell surface. The transfer of resistance trait across various genera occurs through horizontal gene transfer. Two most common setting where horizontal gene transfer occurs is 1. environment which serves as natural reservoir for antimicrobial resistant pathogens and 2. is nosocomial/hospital based.
Previously common treatable bacterial infections such as sepsis, UTIs, sexually transmitted diseases etc. are exhibiting high prevalence of AMR suggesting depletion of treatment options. All these are broadly categorized as ESKAPE pathogens. ESKAPE consists of vancomycin-resistant Enterococcus faecium; methicillin-resistant Staphylococcus aureus; Klebsiella pneumoniae, Acinetobacter baumannii; Pseudomonas aeruginosa, and Enterobacter spp., which are multi-drug resistant, including containing ESBL and resistance to carbapenems. AMR accounts for 15.5% hospital cases with 700,000 annual deaths. Two members of this group i.e. Carbapenem-resistant Acinetobacter and Carbapenem-resistant Enterobacteriaceae has been officially classified in top five antibiotic resistant strains according to 2019 report of CDC. This led to creation of global priority pathogen list by WHO with an objective to prioritize research and treatment. It classifies into 3 categories, critical, high, and medium with 4 ESKAPE pathogens officially designated as critical priority list and 2 as high priority list. Currently both E. coli and K. pneumoniae started exhibiting 4-92% resistance ciprofloxacin creating an alarming situation. It seems the last resort treatments are getting exhausted as organisms like Klebsiella spp., E. coli, Staphlylococcus aureus have started exhibiting resistance carbapenems, colistin and methicillin. In case Mycobacterium, the case seems grimmer. As of 2019, WHO estimates 500,000 ripampin resistant TB cases with incidence of 3% in newer cases and 18% in pan resistant cases.
So, we stand in a conjunction where global regulation authorities like WHO started formulating regulations to curb antibiotic misuse and keeping current treatment practices effective. For effective combat of AMR strains, newer antibiotics discovery and discovery of adjuvants to effectively sensitize the current antibiotics seems more logical. The impetus to this thought was with the discovery of clauvulinic acid which reversed AMR towards beta-lactamases. With almost no new antibiotic discovery program and handful molecules in various clinical trials, it becomes imperative to venture into newer technologies to combat AMR.
Clustered Regularly Interspaced Short Palindromic Repeats or CRISPR seems one of the newer and most promising technologies which can be evaluated for combating AMR. Acquisition of virulence, fitness and antibiotic resistance genes from environment is an important step in bacterial adaptive system to survive. Other mobile genetic elements (MGEs) also get integrated in bacterial genome from bacteriophages too which becomes unnecessary baggage for bacteria. In some cases these phenomenon lead to conversion of avirulent strains to virulent strains e.g. Vibrio cholerae. To overcome this, bacteria posses programmable systems called CRISPR. Their initial discovery was thought to be gene editing tool to fight bacteriophases. But later discovery of specific RNA systems called Cas9 revealed their precise nature.
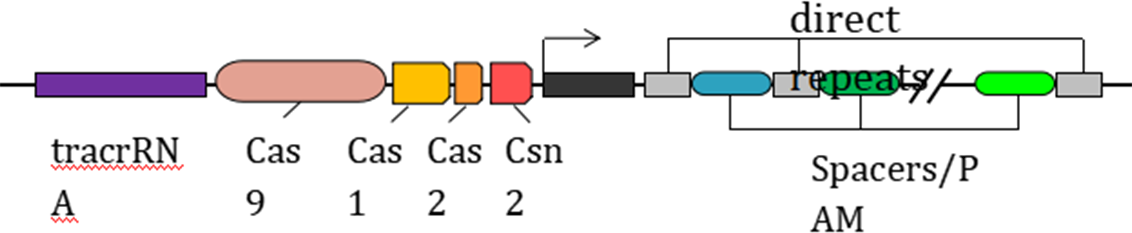
The CRISPR-Cas9 system consists of tracrRNA which recognizes specific sequences, Cas9 RNA and PAM: Protospacer Adjacent Motif. Upon expression Cas9 expresses a nuclease which first binds at PAM and breaks CRISPR-encoded-crRNA of desired sequenced and introducing either point mutation/gene knock down can be achieved. This can be effectively exploited against antibiotic resistant genes to reverse AMR with requirement of the genome of the organism is sequenced. These PAM can be identified in the organism and specific nucleotide sequence can be integrated to target the specific gene. The deliverance system needs to be optimized for introduction into wide host. Similar approaches has been exploited for beta-lactamases, where beta-lactamase gene sequences were cloned upstream of PAM sequences and their introduction led to reversal of bet-lactam resistance.
